Publications
Weakly supervised multi-modal contrastive learning framework for predicting the HER2 scores in breast cancer
Jun Shi, Dongdong Sun, Zhiguo Jiang, Jun Du, Wei Wang, Yushan Zheng*, Wu Haibo*
Computerized Medical Imaging and Graphics, 2025
Pathology report generation from whole slide images with knowledge retrieval and multi-level regional feature selection
Dingyi Hu, Zhiguo Jiang, Jun Shi, Fengying Xie, Kun Wu, Kunming Tang, Ming Cao, Jianguo Huai, Yushan Zheng*
Computer Methods and Programs in Biomedicine, 2025
A deep-learning model for predicting tyrosine kinase inhibitor response from histology in gastrointestinal stromal tumor
Xue Kong#, Jun Shi#, Dongdong Sun#, Lanqing Cheng, Can Wu, Zhiguo Jiang, Yushan Zheng*, Wei Wang*, Haibo Wu*
The Journal of Pathology, 2025
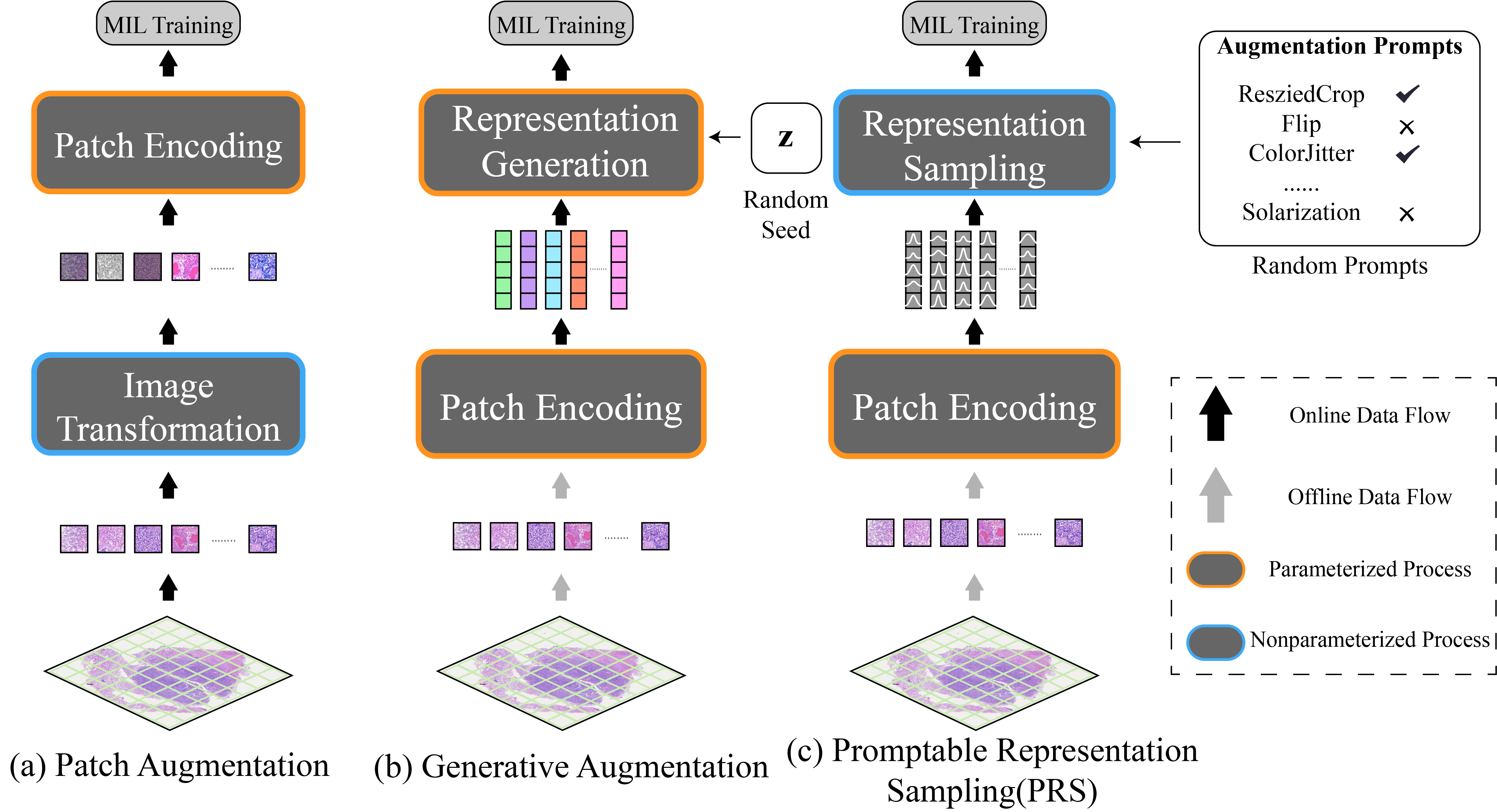
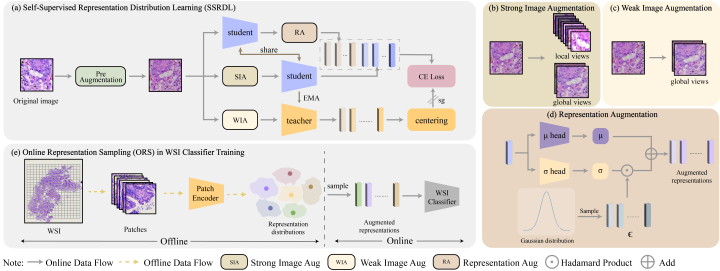
Promptable Representation Distribution Learning and Data Augmentation for Gigapixel Histopathology WSI Analysis
Kunming Tang, Zhiguo Jiang, Jun Shi, Wei Wang, Haibo Wu, Yushan Zheng*
The 39th Annual AAAI Conference on Artificial Intelligence (AAAI), 2025
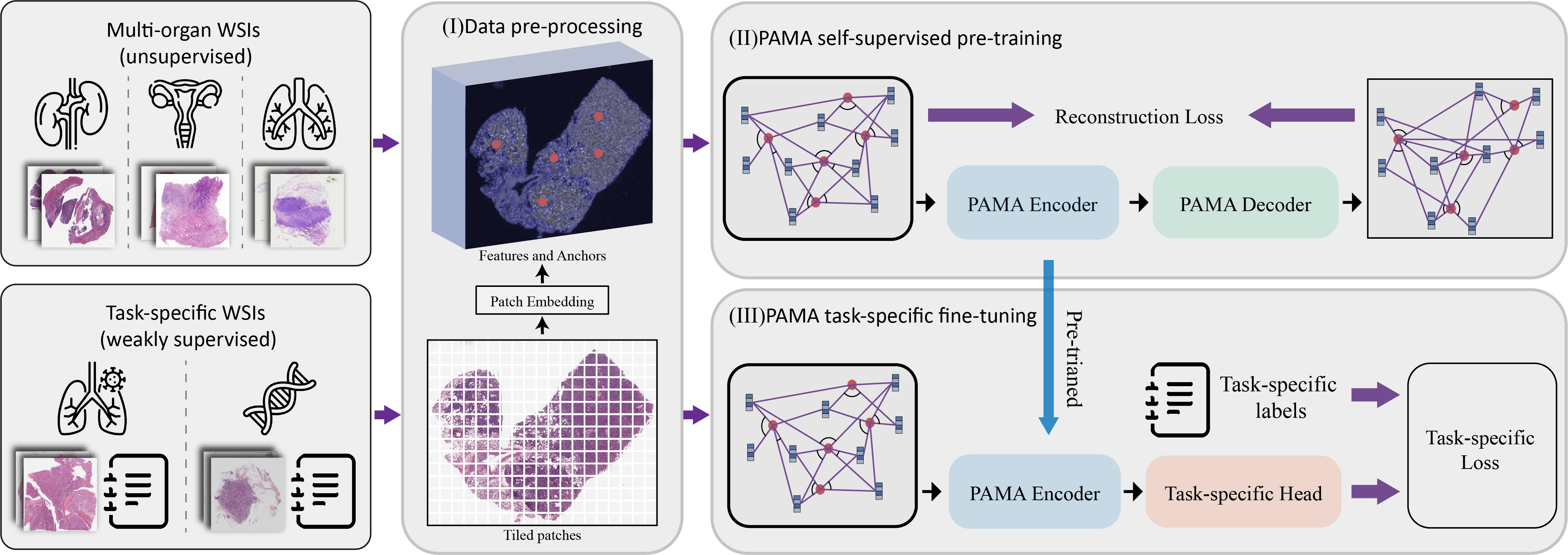
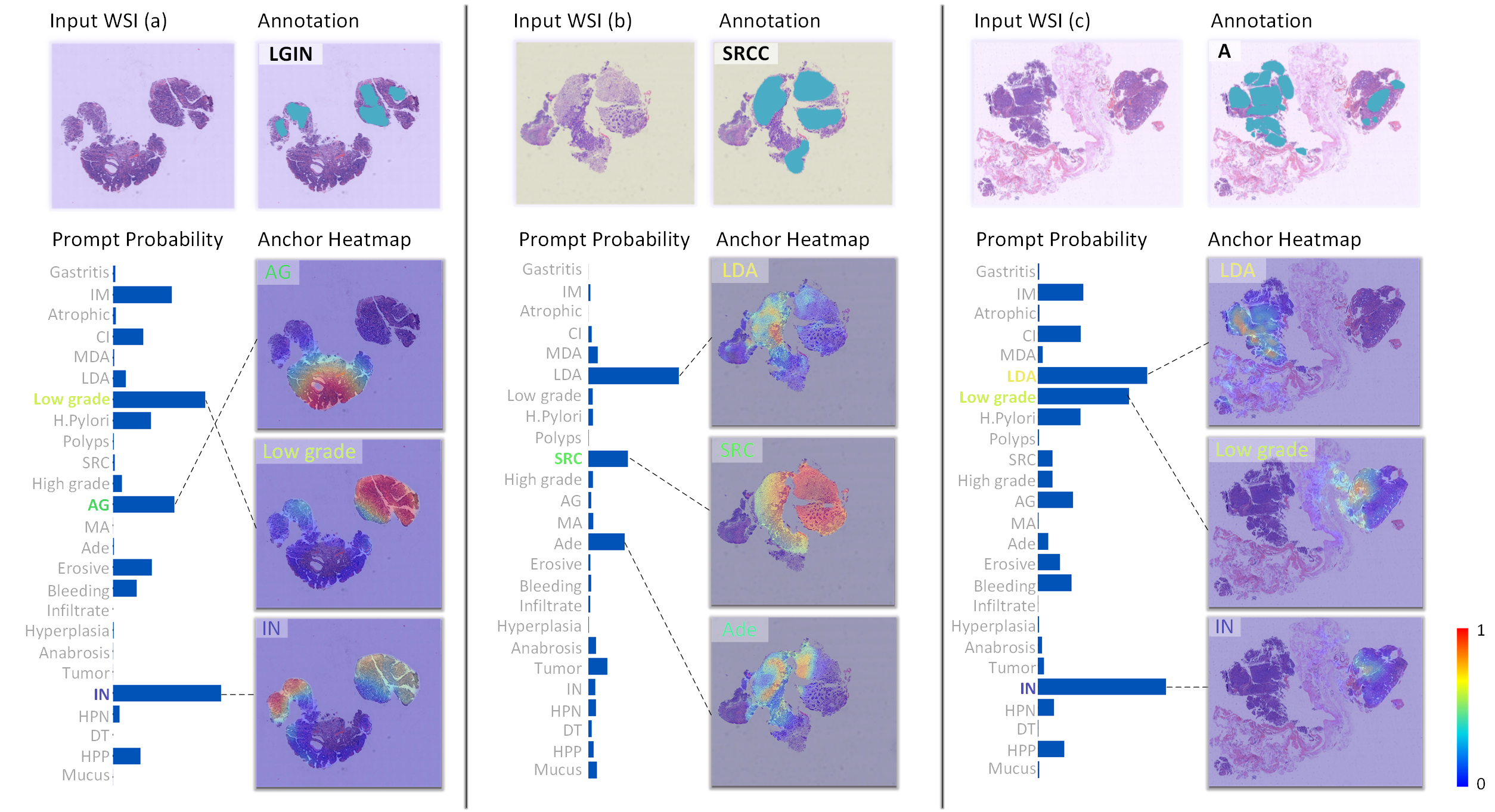
Pan-cancer Histopathology WSI Pre-training with Position-aware Masked Autoencoder
Kun Wu, Zhiguo Jiang, Kunming Tang, Jun Shi, Fengying Xie, Wei Wang, Haibo Wu, Yushan Zheng*
IEEE Transactions on Medical Imaging, 2024
Self-Supervised Representation Distribution Learning for Reliable Data Augmentation in Histopathology WSI Classification
Kunming Tang, Zhiguo Jiang, Kun Wu, Jun Shi, Fengying Xie, Wei Wang, HaiboWu*, Yushan Zheng*
IEEE Transactions on Medical Imaging, 2024
Prediction of Epidermal Growth Factor Receptor Mutation Subtypes in Non--Small Cell Lung Cancer From Hematoxylin and Eosin--Stained Slides Using Deep Learning
Wanqiu Zhang#, Wei Wang#, Yao Xu#, Kun Wu#, Jun Shi, Ming Li, Zhengzhong Feng*, Yinhua Liu*, Yushan Zheng*, Haibo Wu*
Laboratory Investigation, 2024
Report-Guided Cross-Modal Representation Learning for Predicting EGFR Mutations by Whole Slide Image
Qi Qiao, Jun Shi, Zhiguo Jiang, Wei Wang, Haibo Wu, Yushan Zheng*
2024 IEEE International Conference on Bioinformatics and Biomedicine (BIBM), 2024
Partial-Label Contrastive Representation Learning for Fine-grained Biomarkers Prediction from Histopathology Whole Slide Images
Yushan Zheng, Kun Wu, Jun Li, Kunming Tang, Jun Shi, Haibo Wu, Zhiguo Jiang*, and Wei Wang*
IEEE Jounal of Biomedical and Health Informatics, 2024
Leveraging IHC Staining to Prompt HER2 Status Prediction from HE-Stained Histopathology Whole Slide Images
Yuping Wang, Dongdong Sun, Jun Shi, Wei Wang, Zhiguo Jiang, Haibo Wu, Yushan Zheng*
MICCAI Workshop on Machine Learning in Medical Imaging, 2024
Lifelong Histopathology Whole Slide Image Retrieval via Distance Consistency Rehearsal
Xinyu Zhu, Zhiguo Jiang, Kun Wu, Jun Shi, and Yushan Zheng*
Medical Image Computing and Computer Assisted Intervention (MICCAI), 2024
SlideGCD: Slide-based Graph Collaborative Training with Knowledge Distillation for Whole Slide Image Classification
Tong Shu, Jun Shi, Dongdong Sun, Zhiguo Jiang, and Yushan Zheng*
Medical Image Computing and Computer Assisted Intervention (MICCAI), 2024
Masked hypergraph learning for weakly supervised histopathology whole slide image classification
Jun Shi, Tong Shu, Kun Wu, Zhiguo Jiang, Liping Zheng, Wei Wang, Haibo Wu, and Yushan Zheng*
Computer Methods and Programs in Biomedicine, 2024
Genomics-Embedded Histopathology Whole Slide Image Encoding for Data-efficient Survival Prediction
Kun Wu, Zhiguo Jiang, Xinyu Zhu, Jun Shi, and Yushan Zheng*
Medical Imaging with Deep Learning, 2024
Histopathology language-image representation learning for fine-grained digital pathology cross-modal retrieval
Dingyi Hu, Zhiguo Jiang, Jun Shi, Fengying Xie, Kun Wu, Kunming Tang, Ming Cao, Jianguo Huai, and Yushan Zheng*
Medical Image Analysis, 2024
Position-Aware Masked Autoencoder for Histopathology WSI Representation Learning
Kun Wu, Yushan Zheng*, Jun Shi*, Fengying Xie, and Zhiguo Jiang
Medical Image Computing and Computer Assisted Intervention (MICCAI), 2023
A Key-Points Based Anchor-Free Cervical Cell Detector
Tong Shu, Jun Shi*, Yushan Zheng*, Zhiguo Jiang, and Lanlan Yu
IEEE Engineering in Medicine & Biology Society (EMBC), 2023
Cervical Cell Classification Using Multi-Scale Feature Fusion and Channel-Wise Cross-Attention
Jun Shi, Xinyu Zhu, Yuan Zhang, Yushan Zheng*, Zhiguo Jiang, and Liping Zheng
IEEE 20th International Symposium on Biomedical Imaging (ISBI), 2023
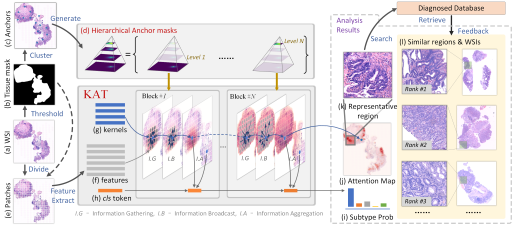
Kernel Attention Transformer for Histopathology Whole Slide Image Analysis and Assistant Cancer Diagnosis
Yushan Zheng, Jun Li, Jun Shi*, Fengying Xie, Jianguo Huai, Ming Cao, and Zhiguo Jiang*
IEEE Transactions on Medical Imaging, 2023
Histopathology Cross-Modal Retrieval based on Dual-Transformer Network
Dingyi Hu, Yushan Zheng*, Fengying Xie, Zhiguo Jiang, and Jun Shi
IEEE International Conference on Bioinformatics and Bioengineering (BIBE), 2022
Kernel Attention Transformer (KAT) for Histopathology Whole Slide Image Classification
Yushan Zheng*, Jun Li, Jun Shi, Fengying Xie, Zhiguo Jiang
Medical Image Computing and Computer Assisted Intervention (MICCAI), 2022
Lesion-Aware Contrastive Representation Learning For Histopathology Whole Slide Images Analysis
Jun Li, Yushan Zheng*, Kun Wu, Jun Shi*, Fengying Xie, Zhiguo Jiang
Medical Image Computing and Computer Assisted Intervention (MICCAI), 2022
Global-local attention network for weakly supervised cervical cytology ROI analysis
Jun Shi*, Kun Wu, Yushan Zheng*, Yuxin He, Jun Li, Zhiguo Jiang and Lanlan Yu
IEEE 19th International Symposium on Biomedical Imaging, 2022
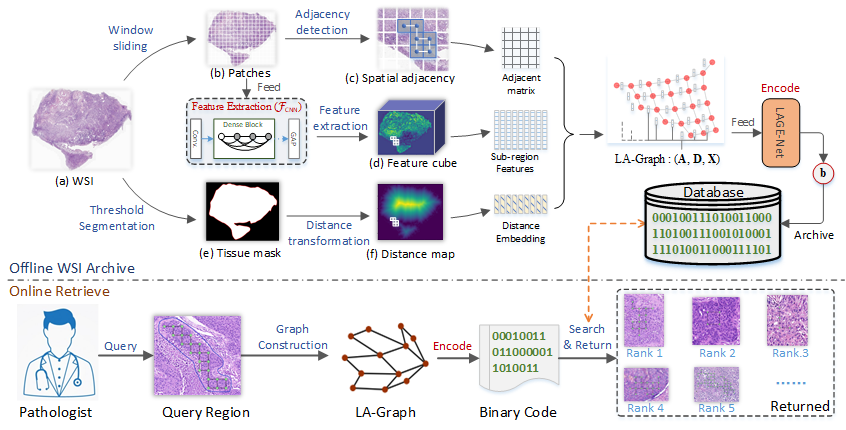
Encoding Histopathology Whole Slide Images with Location-aware Graphs for Diagnostically Relevant Regions Retrieval
Yushan Zheng, Zhiguo Jiang*, Jun Shi*, Fengying Xie, Haopeng Zhang, Wei Luo, Dingyi Hu, Shujiao Sun, Zhongmin Jiang, and Chenghai Xue
Medical Image Analysis, 2022
Histopathological whole slide images (WSIs) are gigapixel images widely used in cancer diagnosis. Each WSI may include multiple diagnostically relevant regions, making them challenging to represent. Meanwhile, the spatial relationships between tissue regions are often diagnostically significant. This paper proposes a novel location-aware graph encoding network (LaGeNet) for representing histopathology WSIs that captures both region-wise feature representations and spatial relationships. Unlike existing multiple-instance learning (MIL) methods that focus solely on region features, our approach builds a location graph from region nodes with positional encoding and creates a diagnostic node for global feature learning. The model is trained end-to-end using a contrastive learning strategy to capture diagnostic similarities between WSIs. We evaluate our approach on five datasets from different organs, demonstrating superior performance in diagnostically-relevant region retrieval compared to state-of-the-art methods. Our LaGeNet framework provides an effective approach for encoding gigapixel histopathology images while preserving spatial information.
Frequency-based convolutional neural network for efficient segmentation of histopathology whole slide images
Wei Luo, Yushan Zheng, Dingyi Hu, Jun Li, Chenghai Xue, and Zhiguo Jiang
International Conference on Image and Graphics (ICIG), 2021
Diagnostic Regions Attention Network (DRA-Net) for Histopathology WSI Recommendation and Retrieval
Yushan Zheng, Zhiguo Jiang*, Jun Shi, Fengying Xie, Haopeng Zhang, Huai Jianguo, Cao Ming, and Yang Xiaomiao
IEEE Transactions on Medical Imaging, 2021
Stain standardization capsule for application-driven histopathological image normalization
Yushan Zheng, Zhiguo Jiang*, Haopeng Zhang, Fengying Xie, Dingyi Hu, Shujiao Sun, Jun Shi, and Chenghai Xue
IEEE Journal of Biomedical and Health Informatics, 2021
Tracing Diagnosis Paths on Histopathology WSIs for Diagnostically Relevant Case Recommendation
Yushan Zheng*, Zhiguo Jiang, Haopeng Zhang, Fengying Xie, and Jun Shi
Medical Image Computing and Computer Assisted Interventions (MICCAI), 2020
Informative retrieval framework for histopathology whole slides images based on deep hashing network
Dingyi Hu, Yushan Zheng*, Haopeng Zhang, Jun Shi, Fengying Xie, and Zhiguo Jiang
IEEE 17th International Symposium on Biomedical Imaging (ISBI), 2020
Cancer Sensitive Cascaded Networks (CSC-Net) for Efficient Histopathology Whole Slide Image Segmentation
Shujiao Sun, Huining Yuan, Yushan Zheng*, Haopeng Zhang, Dingyi Hu, and Zhiguo Jiang
IEEE 17th International Symposium on Biomedical Imaging (ISBI), 2020
Stain Standardization Capsule: A pre-processing module for histopathological image analysis
Yushan Zheng*, Zhiguo Jiang, Haopeng Zhang, Jun Shi, and Fengying Xie
MICCAI Workshop -- Computational Pathology (COMPAY19)
Encoding histopathological WSIs using GNN for scalable diagnostically relevant regions retrieval
Yushan Zheng*, Bonan Jiang, Jun Shi, Haopeng Zhang, and Fengying Xie
Medical Image Computing and Computer Assisted Interventions (MICCAI), 2019
A Comparative Study of CNN and FCN for Histopathology Whole Slide Image Analysis
Shujiao Sun, Bonan Jiang, Yushan Zheng*, and Fengying Xie
International Conference on Image and Graphics (ICIG), 2019
Adaptive Color Deconvolution for Histological WSI Normalization
Yushan Zheng, Zhiguo Jiang*, Haopeng Zhang, Fengying Xie, Jun Shi and Chenghai Xue
Computer Methods and Programs in Biomedicine, 2019
Histopathological Whole Slide Image Analysis Using Context-based CBIR
Yushan Zheng, Zhiguo Jiang, Haopeng Zhang*, Fengying Xie, Yibing Ma, Huaqiang Shi and Yu Zhao
IEEE Transactions on Medical Imaging, 2018
Size-scalable Content-based Histopathological Image Retrieval from Database that Consists of WSIs
Yushan Zheng, Zhiguo Jiang*, Haopeng Zhang, Fengying Xie, Yibing Ma, Huaqiang Shi and Yu Zhao
IEEE Journal of Biomedical and Health Informatics, 2018
Feature Extraction from Histopathological Images Based on Nucleus-guided Convolutional Neural Network for Breast Lesion Classification
Yushan Zheng, Zhiguo Jiang, Fengying Xie*, Haopeng Zhang, Yibing Ma, Huaqiang Shi and Yu Zhao
Pattern Recognition, 2017
Content-based Histopathological Image Retrieval for Whole Slide Image Database Using Binary Codes
Yushan Zheng*, Zhiguo Jiang, Yibing Ma, Haopeng Zhang, Fengying Xie, Huaqiang Shi and Yu Zhao
SPIE Medical Imaging, 2017
Retrieval of Pathology Image for Breast Cancer Using PLSA Model Based on Texture and Pathological Features
Yushan Zheng*, Zhiguo Jiang, Jun Shi and Yibing Ma
IEEE International Conference on Image Processing (ICIP), 2014